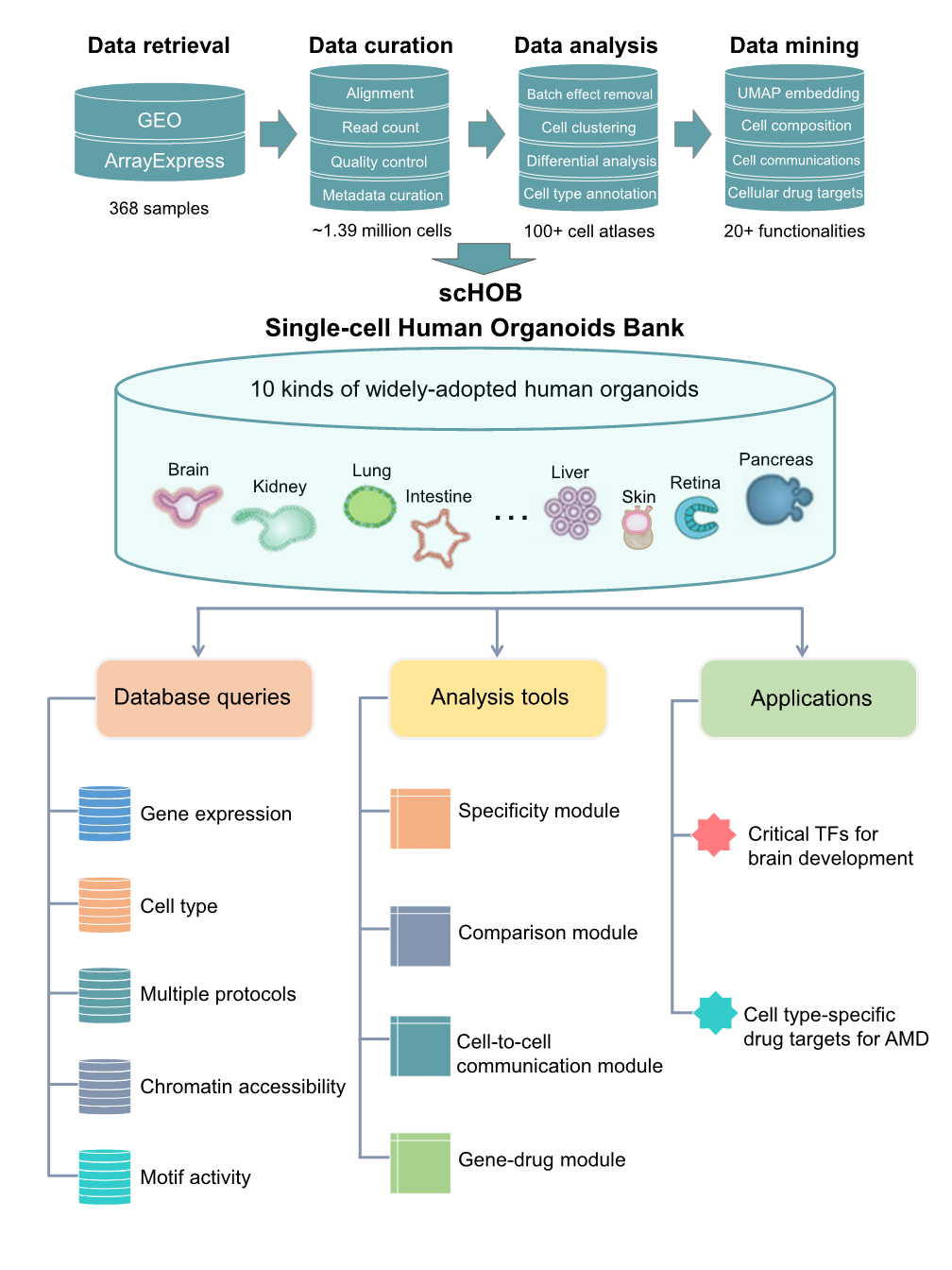
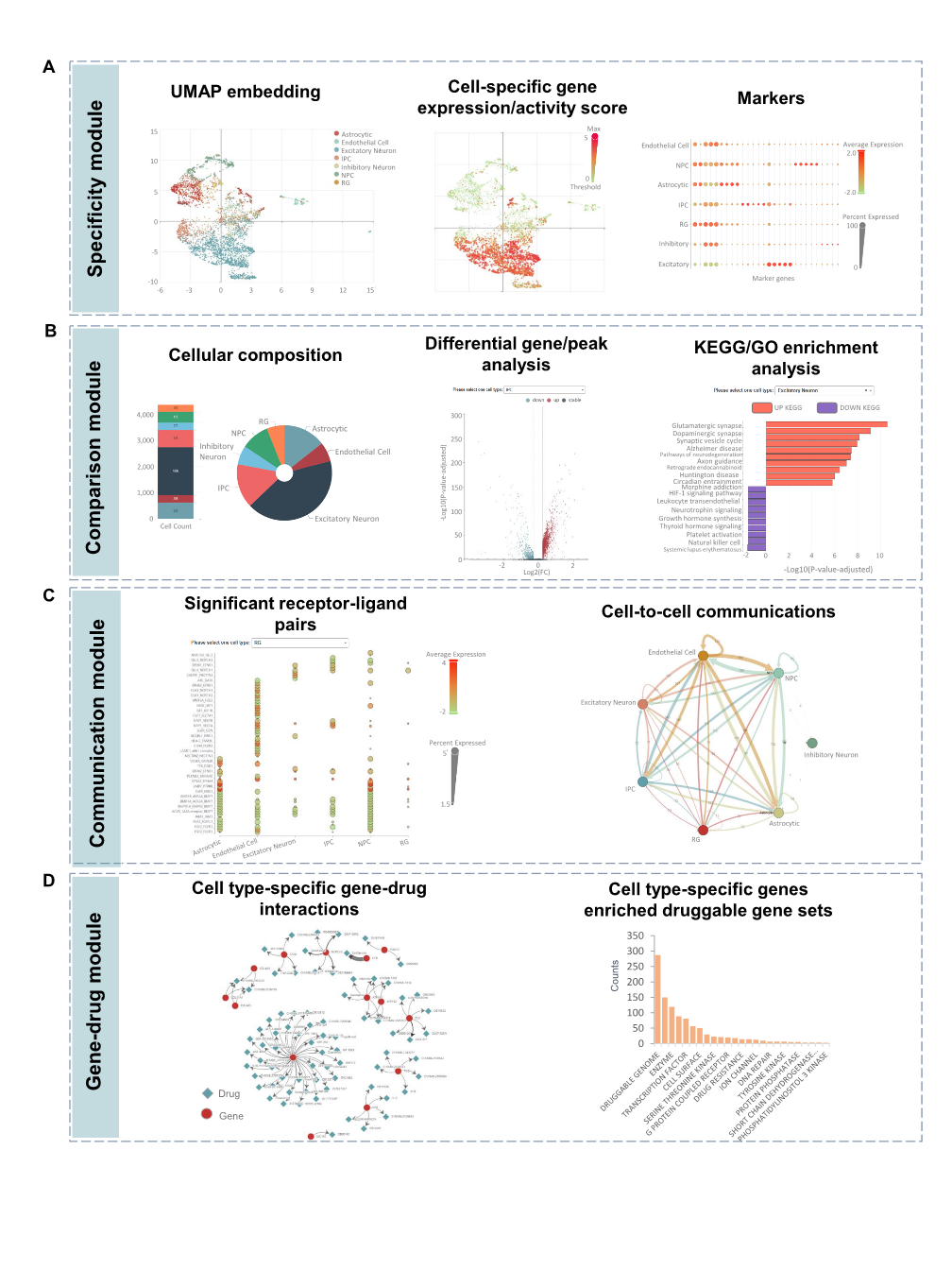
Major depressive disorder (MDD) is a heritable mental illness affecting over 300 million people worldwide. Despite numerous genetic variants being identified, many are located in non-coding regions, complicating their functional interpretation. Identifying trait-relevant cell populations using single-cell data is essential for investigating the genetic mechanisms underlying MDD. Here, we introduce scDepBrain (single-cell Depression Brain map), a resource that integrates brain single-cell transcriptomic profiles (5,049,376 cells from 1,599 brain samples) with large-scale GWAS data (500,199 samples) to identify MDD-relevant cell types and subpopulations.
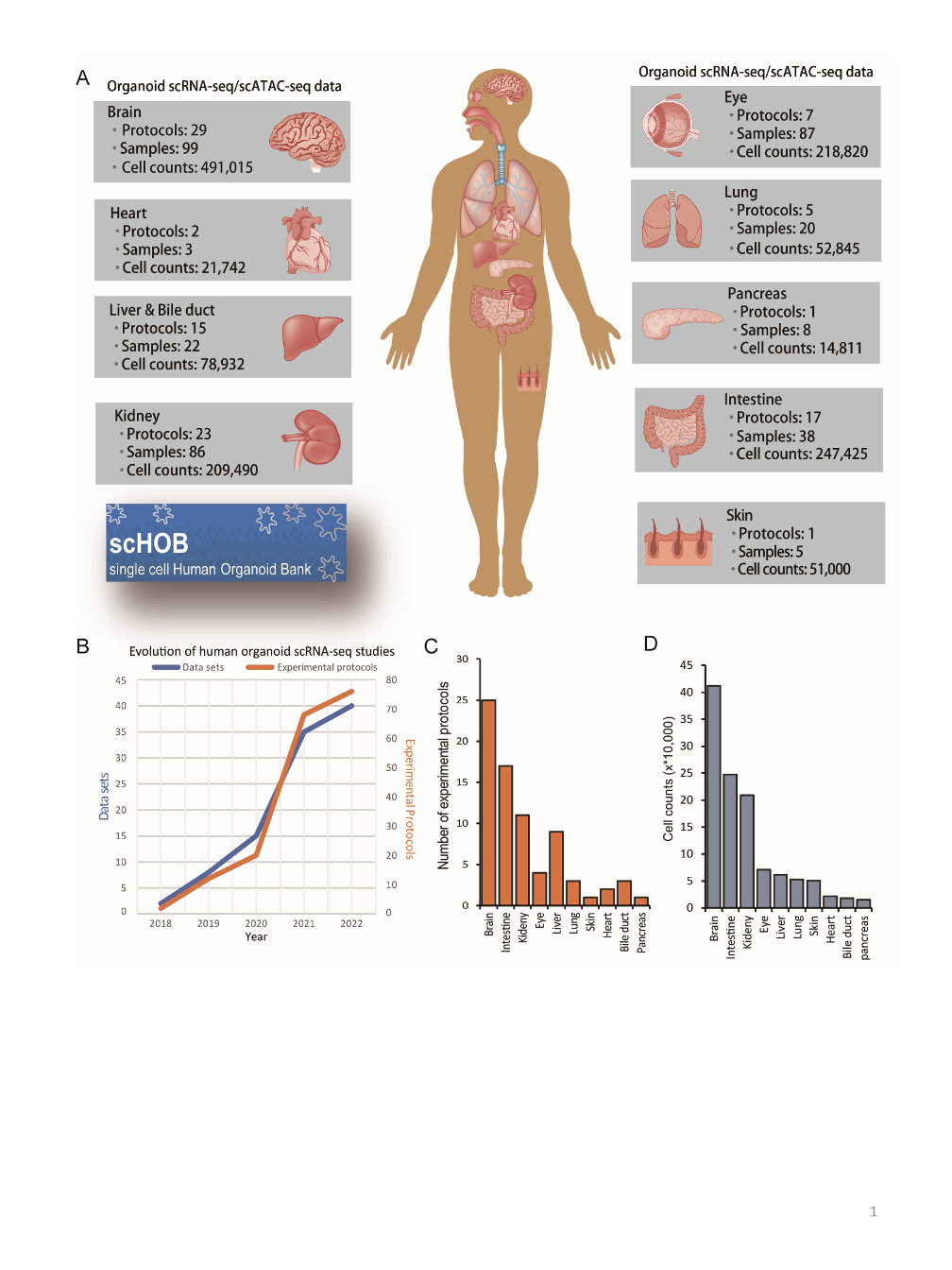
We curated and analyzed brain single-cell and single-nuclei sequencing data from 34 published studies, categorizing them into one discovery dataset and five replication datasets based on species origin (human, mouse, or organoid), brain developmental stage (adult or developing), and health status (disease or healthy): Discovery dataset: 327,639 cells from 140 healthy human brain samples across 14 anatomical regions. Replication datasets: (1) Human adult brain: 3,369,219 cells from 606 healthy brain samples across 10 regions. (2) Human developing brain: 599,221 cells from 733 brain samples, covering gestational weeks 6 to 40 and up to 8 months postnatal. (3) Mouse brain: 197,026 cells from 23 samples across 5 regions. (4) Human brain organoid: 396,049 cells from 25 organoid samples. (5) Human MDD brain: 160,222 cells from 37 MDD patients and 34 healthy controls. We applied four widely used integration methods—scPagwas, scDRS, LDSC-SEG, and MAGMA_CellTyping—to pinpoint MDD-relevant cell types and subpopulations. The resulting data are freely available for search, visualization, and download.
Disclaimer: This resource is intended for purely research purposes. It should not be used for emergencies or medical or professional advice.